I got to thinking about something and ran a few tests. The crown jewel for me in the malt data that I have is A.J.'s Weyermann Floor Pils:
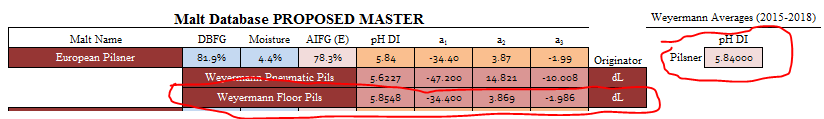
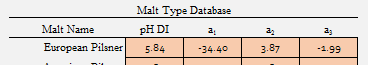
It has all 3 terms and a pH DI that corresponds to past and present pH DI values from lot analysis from Weyermann. I merely took it, changed the pH DI to my Weyermann average from my lot analysis data bank, checked the 2019 analysis values across their pilsner range, and used it for my European Pilsner malt class:
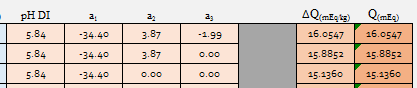
I want to see the effect of simply taking this and removing a3 and a2 sequentially. My thought being that anyone can stop at any point in the process of tritrating malt and simply not go beyond a1.
I used the "Custom Malt Parameters" slots and just forced it to be a dual and single point set:
I set source minerals to zero with Alkalinity set to 1.58 mg/l as CaCO3, remembering that value was what A.J. always spec'd for DI water. 1 Kg each of European Pilsner, Custom 1 and Custom 2 yields:
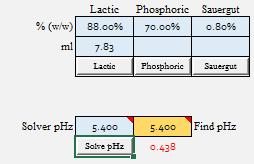
After just whipping up some reasonable batch parameters (32 l strike volume, 5.67 kg of malt, DI water, no added minerals for simplicity), I ran each malt to see what value of 88% Lactic Acid it would take to go from Grist pH to 5.4:
| European Pilsner (3 Term): |
| European Pilsner (2 Term): |
|
| European Pilsner (1 Term): |
|
Grist pH = 5.838
| Grist pH = 5.838
| Grist pH = 5.838
|
Lactic Acid to 5.4 = 7.91 ml
| Lactic Acid to 5.4 = 7.83 ml
| Lactic Acid to 5.4 = 7.46 ml
|
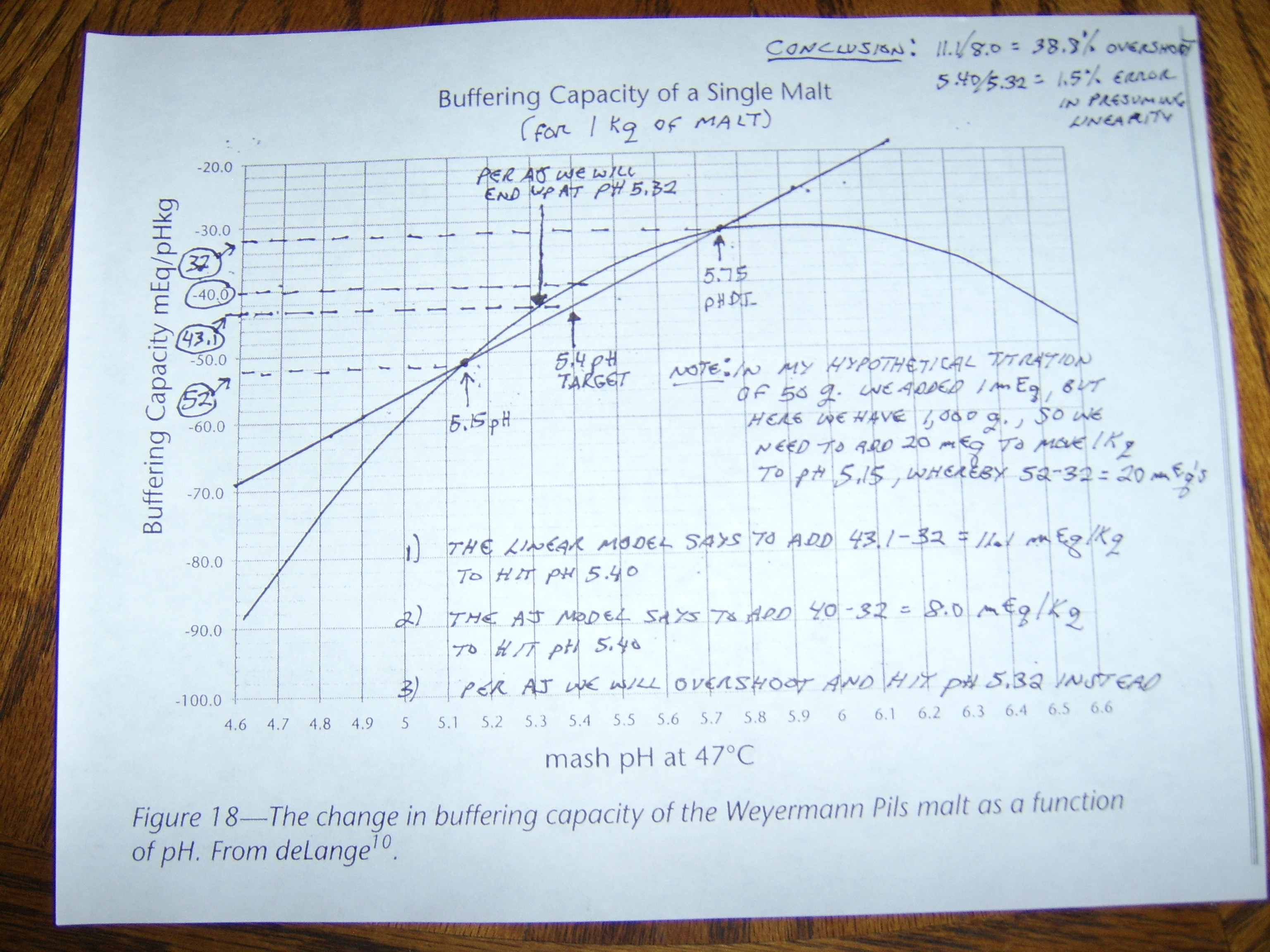
I'm not exactly sure what my point is other than if we have a reliable data set for a malt, that makes sense from a practical standpoint, we can show that we really do not seem to introduce much error in the amount of acid required to acidify the batch from assuming it to be linear.
The rub here is that we can only test this if we have a 3 point data set. I know that Riffe was confident in the point I just made when he normalized 3 point data sets to linear, as it agreed pretty much with what I have shown. However, we can never really be sure that single titration point data is as "accurate" for crystal mats, roasted malts, adjuncts, etc. because we don't have any 2 or 3 point data sets to compare them to.




















































![Craft A Brew - Safale S-04 Dry Yeast - Fermentis - English Ale Dry Yeast - For English and American Ales and Hard Apple Ciders - Ingredients for Home Brewing - Beer Making Supplies - [1 Pack]](https://m.media-amazon.com/images/I/41fVGNh6JfL._SL500_.jpg)