Owly055
Well-Known Member
- Joined
- Feb 28, 2014
- Messages
- 3,008
- Reaction score
- 686
I found this too interesting not to share. I've had a passion for microbes and microbial cultures of many kinds since childhood. A fascinating world. Yeast is a fungus, and it appears that G Candidum bridges the gap between conventional yeasts and fungi. Enjoy ;-)
http://microbialfoods.org/geotrichum-candidum-mold-transition/
Geotrichum candidum: A yeast holding on to its moldy past
23/12/15 | Posted in cheese & dairy by Benjamin Wolfe
Have you ever noticed those goat’s milk cheeses with the wrinkly surface at the cheese shop? They look like a fuzzy white brain or a dusty grey coral and they smell like sweet, buttery flatulence. Those aesthetics and aromas come from the growth of the fungus Geotrichum candidum. Using in-depth genomic sequencing, French scientists recently unlocked the evolutionary history of this important cheese microbe and revealed a fungus with an identity crisis.
All true yeasts are descendants of molds; they evolved a single-celled lifestyle from their moldy multi-cellular ancestors. Throughout this evolutionary process, they let go of habits such as growth using the filaments that create the fuzzy appearance of most molds. For a long time, Geotrichum candidum (hereafter G. candidum) has been a mycological mystery to both cheese makers and scientists. It has a fuzzy appearance when it grows on the surface of cheeses or on Petri dishes in the lab, but when you look under the microscope, it also has the appearance of tiny single cells that look like a yeast (see figure below). And when scientists stitched together a family tree of the yeasts based on small segments of DNA sequences, they noticed that G. candidum always clustered with true yeasts like the baker’s yeast Saccharomyces cerevisiae, suggesting that it was a yeast with moldy tendencies. Was this an example of a yeast that only partly transitioned from its moldy past to a yeasty lifestyle? Why had it held on to some previous mold traits while other yeasts did not?
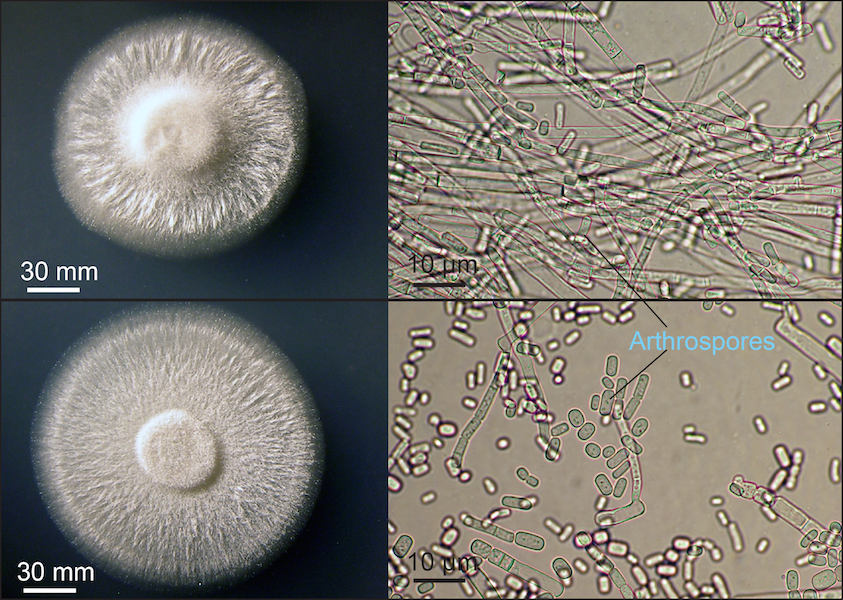
Variation in macro- and microscopic appearance of Geotrichum candidum. Photos on left show colonies growing on Peri dishes in the lab. Photos on the right showing cells were taken using a microscope at 400X magnification. Top two photos show an example of a mold-like strain. Bottom two photos show an example of a yeast-like strain.
To better understand this mold in transition, a team of French scientists recently sequenced and analyzed the entire genome of G. candidum. This whole genome sequence of its DNA gives a genetic blueprint of the fungus. It identifies all the genes that determine traits like its ability to grow on cheese and how it appears when it grows. In an approach called comparative genomics, they then compared this genetic blueprint to similar blueprints that had already been generated for true single-celled yeasts like Saccharomyces cerevisiae, Yarrowia lipolytica, and Debaryomyces hansenii as well as molds such as the plant pathogen Sclerotinia sclerotiorum and the soil fungus Aspergillus nidulans. By determining which genes Geotrichum has relative to these yeasts and molds, they determined which mold genes G. candidum had retained and which unique yeast genes it had evolved. They could also determine if there were any unique genes that might explain why G. candidum grows so well on cheese.
Results from comparing the genetic blueprints revealed some important clues as to why G. candidum is such a fuzzy yeast. Chitin is a chemical compound that is major building block of fungal cell walls. Interestingly, G. candidum has many more genes involved with the production of chitin than other yeasts. Because chitin is important in building the filaments that make up a mold, this may be one reason G. candidum often produces filaments and appears like a mold.
Other genes detected in the G. candidum blueprint highlight its propensity to live on plants. We often think of this as a dairy-loving yeast because it is so widely used in cheese making, but strains of G. candidum are often found on plants and in soils where dead plant material is abundant. Many molds use enzymes called cellulases to decompose cellulose, an important building material in plant cells. During the decomposition process they convert cellulose into sugars that they can use to fuel their own growth. Most yeasts have lost these cellulase genes because they no longer depend on decomposition of plants as a means of survival. But G. candidum has a suite of cellulases that are common in mold genomes that it can use to break down cellulose in the environment. This finding helps explain the ecological versatility of G. candidum and its ability to grow on plants.
In the cheese environment, G. candidum produces many lipases, enzymes that break down the fats in cheese. Surprisingly, the scientists found a particular type of lipase gene that was abundant in the genome of G. candidum but absent from all other yeast genomes. They then determined that all of these lipase genes are found in the mold genomes. This suggests that the ability to be such a prolific producer of useful enzymes is a trait that G. candidum has retained from its moldy ancestors.

A closeup view of the wrinkles on the surface of two cheeses created by the fungus Geotrichum candidum. Photos by Adam Detour.
There are many more interesting findings from this pioneering work. 141 genes that were present in G. candidum were found to be absent from others yeasts, but present in mold genomes. Many of these genes have unknown functions because they haven’t been studied before or are potentially brand new types of genes. By identifying these genes with large genomic datasets, researchers have started to define what it means to be a yeast versus a mold, creating exciting new opportunities to understand the genetic basis of the diversity of fungi.
What might this huge amount of genomic data mean in a practical sense for cheese producers? This data provides some of the first evidence of why the G. candidum strains used in cheesemaking have their specific traits. It might also help us identify other unexplored yeasts that aren’t currently used in the production of cheese or other fermented foods, but might have desirable traits for these microbial foods. In some applications, it might be useful to have a yeast that has held on to some aspects of its moldy origins. We can now use these genomic databases to find those yeasts. We might also be able to use these data to improve yeasts through classic breeding approaches. By mating together different strains or even species of yeasts, just as crop breeders cross different varieties and species of plants, we might be able to make hybrids that provide novel flavors or aesthetics in microbial foods.
For more details on this study, please check out the full article here:
Morel, Guillaume, et al. “Differential gene retention as an evolutionary mechanism to generate biodiversity and adaptation in yeasts.” Scientific Reports 5 (2015). http://www.nature.com/articles/srep11571?WT.ec_id=SREP-631-20150630#abstract
Post written by Benjamin Wolfe.
http://microbialfoods.org/geotrichum-candidum-mold-transition/
Geotrichum candidum: A yeast holding on to its moldy past
23/12/15 | Posted in cheese & dairy by Benjamin Wolfe
Have you ever noticed those goat’s milk cheeses with the wrinkly surface at the cheese shop? They look like a fuzzy white brain or a dusty grey coral and they smell like sweet, buttery flatulence. Those aesthetics and aromas come from the growth of the fungus Geotrichum candidum. Using in-depth genomic sequencing, French scientists recently unlocked the evolutionary history of this important cheese microbe and revealed a fungus with an identity crisis.
All true yeasts are descendants of molds; they evolved a single-celled lifestyle from their moldy multi-cellular ancestors. Throughout this evolutionary process, they let go of habits such as growth using the filaments that create the fuzzy appearance of most molds. For a long time, Geotrichum candidum (hereafter G. candidum) has been a mycological mystery to both cheese makers and scientists. It has a fuzzy appearance when it grows on the surface of cheeses or on Petri dishes in the lab, but when you look under the microscope, it also has the appearance of tiny single cells that look like a yeast (see figure below). And when scientists stitched together a family tree of the yeasts based on small segments of DNA sequences, they noticed that G. candidum always clustered with true yeasts like the baker’s yeast Saccharomyces cerevisiae, suggesting that it was a yeast with moldy tendencies. Was this an example of a yeast that only partly transitioned from its moldy past to a yeasty lifestyle? Why had it held on to some previous mold traits while other yeasts did not?

Variation in macro- and microscopic appearance of Geotrichum candidum. Photos on left show colonies growing on Peri dishes in the lab. Photos on the right showing cells were taken using a microscope at 400X magnification. Top two photos show an example of a mold-like strain. Bottom two photos show an example of a yeast-like strain.
To better understand this mold in transition, a team of French scientists recently sequenced and analyzed the entire genome of G. candidum. This whole genome sequence of its DNA gives a genetic blueprint of the fungus. It identifies all the genes that determine traits like its ability to grow on cheese and how it appears when it grows. In an approach called comparative genomics, they then compared this genetic blueprint to similar blueprints that had already been generated for true single-celled yeasts like Saccharomyces cerevisiae, Yarrowia lipolytica, and Debaryomyces hansenii as well as molds such as the plant pathogen Sclerotinia sclerotiorum and the soil fungus Aspergillus nidulans. By determining which genes Geotrichum has relative to these yeasts and molds, they determined which mold genes G. candidum had retained and which unique yeast genes it had evolved. They could also determine if there were any unique genes that might explain why G. candidum grows so well on cheese.
Results from comparing the genetic blueprints revealed some important clues as to why G. candidum is such a fuzzy yeast. Chitin is a chemical compound that is major building block of fungal cell walls. Interestingly, G. candidum has many more genes involved with the production of chitin than other yeasts. Because chitin is important in building the filaments that make up a mold, this may be one reason G. candidum often produces filaments and appears like a mold.
Other genes detected in the G. candidum blueprint highlight its propensity to live on plants. We often think of this as a dairy-loving yeast because it is so widely used in cheese making, but strains of G. candidum are often found on plants and in soils where dead plant material is abundant. Many molds use enzymes called cellulases to decompose cellulose, an important building material in plant cells. During the decomposition process they convert cellulose into sugars that they can use to fuel their own growth. Most yeasts have lost these cellulase genes because they no longer depend on decomposition of plants as a means of survival. But G. candidum has a suite of cellulases that are common in mold genomes that it can use to break down cellulose in the environment. This finding helps explain the ecological versatility of G. candidum and its ability to grow on plants.
In the cheese environment, G. candidum produces many lipases, enzymes that break down the fats in cheese. Surprisingly, the scientists found a particular type of lipase gene that was abundant in the genome of G. candidum but absent from all other yeast genomes. They then determined that all of these lipase genes are found in the mold genomes. This suggests that the ability to be such a prolific producer of useful enzymes is a trait that G. candidum has retained from its moldy ancestors.

A closeup view of the wrinkles on the surface of two cheeses created by the fungus Geotrichum candidum. Photos by Adam Detour.
There are many more interesting findings from this pioneering work. 141 genes that were present in G. candidum were found to be absent from others yeasts, but present in mold genomes. Many of these genes have unknown functions because they haven’t been studied before or are potentially brand new types of genes. By identifying these genes with large genomic datasets, researchers have started to define what it means to be a yeast versus a mold, creating exciting new opportunities to understand the genetic basis of the diversity of fungi.
What might this huge amount of genomic data mean in a practical sense for cheese producers? This data provides some of the first evidence of why the G. candidum strains used in cheesemaking have their specific traits. It might also help us identify other unexplored yeasts that aren’t currently used in the production of cheese or other fermented foods, but might have desirable traits for these microbial foods. In some applications, it might be useful to have a yeast that has held on to some aspects of its moldy origins. We can now use these genomic databases to find those yeasts. We might also be able to use these data to improve yeasts through classic breeding approaches. By mating together different strains or even species of yeasts, just as crop breeders cross different varieties and species of plants, we might be able to make hybrids that provide novel flavors or aesthetics in microbial foods.
For more details on this study, please check out the full article here:
Morel, Guillaume, et al. “Differential gene retention as an evolutionary mechanism to generate biodiversity and adaptation in yeasts.” Scientific Reports 5 (2015). http://www.nature.com/articles/srep11571?WT.ec_id=SREP-631-20150630#abstract
Post written by Benjamin Wolfe.


