I remove a small sample of the 400ml thin slurry. Make a set of serial dilutions by adding 9ml of DI water to a few test tubes then 1ml of the slurry, then 1ml of the mix, etc eventually getting 1:10, 1:100 and 1:1000 samples. The 1:10 factor is what I used to count.
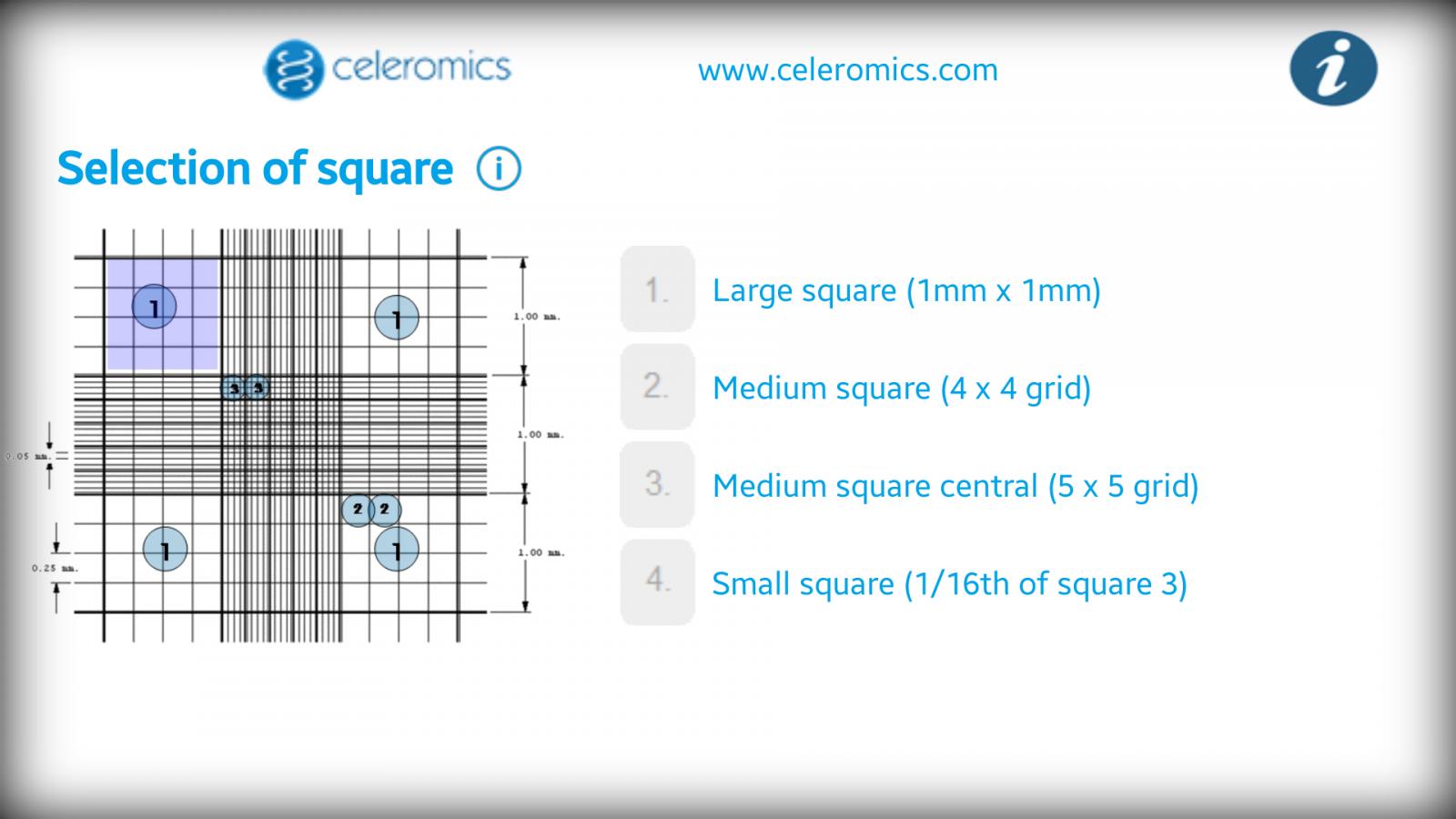
Using my hemocytometer, I count 170 cells in the 16 small (1/16th) squares. Phone app gives me 4.25e7 or 42.5 million cells per ml (1:10 dilution).
Then I add 1ml alkaline methyl violet solution to a 1ml sample of the 1:10 dilution creating a 1:20 dilution that I can assess for viability.
Again using the hemocytometer, I count 92 cells in 16 squares but this time with 9 staining purple (that weren't budding). Phone app gives me 2.30e7 or 23 million cells per ml (1:20 dilution). (92-9)/92x100= 90% viability.
Sooo the average pitch rate between the two counts without dilution factor is 442 million cells per ml. I have 400 ml of slurry. Multiply them together to get a pitch of 176.8 billion cells....of which 90% are viable, so 159 billion viable cells pitched, slightly under the MrMalty calculated rate.
Does this look correct? I hate math.