I think i'm using the wrong terms but in general we are on the same page..
In terms of the actual algorithm we are closer to being on the same page than one might at first think. We must find the pH at which the sum of the proton deficits of all the mash components is 0.
A + B + C + D = 0
A: Total proton deficit of the malts. The proton deficit of malt number i per unit mass is
a1i*(pH - pHdi) + a2i*(pH - pHdi)^2 + a3i*(pH - pHdi)^3
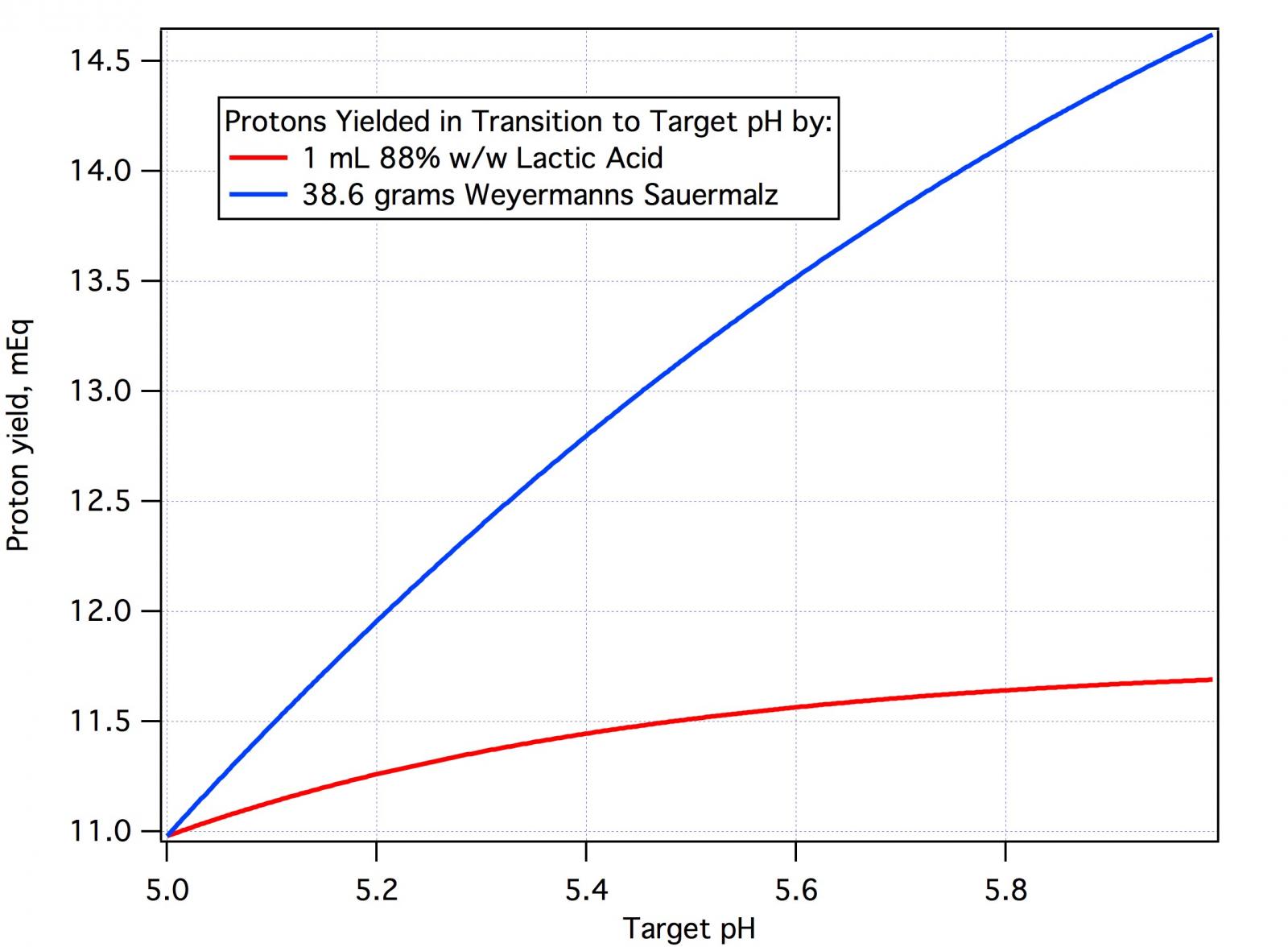
in which pH is the mash pH and pHd the distilled water mash pH. The a1 coefficient dominates the others and is always negative. Thus to get malt i to any pH less than pHdi we have a positive number (e.g. -40*(5.5 - 5.8) = +12). We need protons. A proton deficit is positive. Conversely, if pHdi is less than the target (e.g. -40*(5.5 - 5.3) = -8) we get a negative number. A negative deficit value represents a surfeit of protons from this malt.
For the whole mash we need the proton deficits of all the malts
A = ∑mi*(a1i*(pH - pHdi) + a2i*(pH - pHdi)^2 + a3i*(pH - pHdi)^3 )
where mi is the mass of malt i.
B: B is the proton deficit of the water. It is found from the alkalinity as
B = Vw*Ct*( Q(pH) - Q(pHs) ) in which Vw is the volume of the mash water, Ct is the molar sum of all the carbonic, bicarbonate and carbonate ions in a liter, Q(pH) is the charge on a mole of carbo at the target pH and Q(pHs) is the charge on a mole of carbo at the pH at which the water comes to us. Earlier posts dicuss how to calculate Ct and the two Q's from the alkalinity, the sample pH and the end point pH for the titration which was used to determine the alkalinity. As the target pH is usually lower than the sample pH (pHs) B is usually a positive number representing a proton deficit.
C: C is the proton deficit resulting from the action of calcium and magnesium on phytin in the malt.
C = -Vw*( [Ca++] + [Mg++]/2)/f
in which Vw is, as before, the volume of the mash water (L), the quantities in brackets the calcium and magneisum hardnesses (in mEq/L) and f an unknown factor. Kolbach found f=3.5 for knockout wort. We are sure that it is greater than 3.5 for mash and suspect it is about 7. As C is a negative number it represents a proton surfeit i.e. the reaction supplies hydrogen ions.
D: D is a term which represents the sum of the proton deficits of any acids or bases added to the mash.
D = ∑Vj*Nj
where Vj is the added volume (mL) of solution of added acid or base and Nj is the normality of that solution (the number of mEq of protons absorbed or released by 1 mL of the solution). Nj is positive for bases and negative for acids. As we have seen in earlier posts Nj is a function of pH for lactic and phosphoric acids but not for strong acids like sulfuric and hydrochloric or strong bases (calcium hydroxide, sodium hydroxide, sodium bicarbonate, the latter of which is strong relative to mash pH).
Now we re-write the basic equation as
A(pH) + B(pH) + C + D(pH) = 0
All of the terms, except C, are dependent on pH. Thus, while it is easy to build a spreadsheet that calculates the sum and manipulate the mi and Vj until the sum is 0 in order to design a mash with a particular pH it is not, ostensibly, so easy to find the value of pH which, for a given set of mi and Vj zeroes the sum because A, B and D are not linear in pH. In fact it is quite easy to do this exact thing using Excels Solver but for some reason people are afraid to use that powerful tool. Thus we seek approximations which might allow solution for pH in closed form or, IOW, we seek to linearize each of the terms.
Clearly A is linear in pH if we ignore the squared and cubed terms so that A becomes
A = ∑mi*ai*(pH - pHdi) = ∑mi*ai*pH - ∑mi*ai*pHdi = pH*∑mi*ai - ∑mi*ai*pHdi
We have noticed in running the full algorithm against many problems that
B ≈Vw*0.9*alkalinity
where alkalinity is in units of mEq/L
This eliminates pH dependence from this term.
C does not depend on pH
Lactic acid is almost a strong acid as it is nearly 100% dissociated at mash pH. One mEq of phosphoric acid yields up approximately 1 mEq of protons at pH values typical for mash, sodium bicarbonate absorbs about 1 mEq of protons per mmol when the target is in the usual mash pH range. Thus if we simply change D to
D = ∑VjMj
i.e. substitute the molarity if the acid solutions for the normality we remove the pH dependence and we can write the equation as
pH*∑mi*ai - ∑mi*ai*pHdi + B + C + D = 0
This is easily solved for pH as
pH = (∑mi*ai*pHdi - B - C - D)/∑mi*ai
Now lets make the further assumption that all the ai are about the same and have value a. Then we have
pH = a*∑mi*pHdi/a*∑mi - (B +C + D)/a*∑mi
the a's cancel and this gives
pH = ∑mi*pHdi/∑mi - (B +C + D)/a*∑mi
Note that the first term is simply the weighted average of the individual malt DI mash pH's. B, C and D are the proton deficits of water, calcium (effective) and added acids and bases and a*∑mi is an approximation to the buffering capacity of the entire mash.
The similarity between this and the RPIScotty formula is clear.
The thing about the above that would, to me, at first appear most disturbing, is the replacement of each ai, which can range from -30 to -80, but is usually between say 40 and 60, by a single number. I recall now that the last time this discussion came up I did some sensitivity analysis and found mash pH predictions to be much more dependent on the accuracy of DI mash pH input than on accuracy of ai input. This supports the observation that using actual DI mash pH values, if you have them, is very beneficial (as compared to guessing them from a malt color correlation, for example).












































![Craft A Brew - Safale S-04 Dry Yeast - Fermentis - English Ale Dry Yeast - For English and American Ales and Hard Apple Ciders - Ingredients for Home Brewing - Beer Making Supplies - [1 Pack]](https://m.media-amazon.com/images/I/41fVGNh6JfL._SL500_.jpg)












