Ok after about 1.5 weeks of capturing some local bugs I have a fairly thick layer on the bottom of my mason jar pretty comprable to washing yeast. The smell is not at all unpleasant it almost smells like plain yogurt very lactic and somewhat sweet smelling initially somewhat honey like though there was only Dme and water. What I found interesting was that there were no bubbles like in a regular fermentation, does this mean I didn't catch any yeast that all i got was bacteria ? should I put it back outside ?
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
Howto: Capture Wild Yeast
- Thread starter ericd
- Start date

Help Support Homebrew Talk - Beer, Wine, Mead, & Cider Brewing Discussion Forum:
This site may earn a commission from merchant affiliate
links, including eBay, Amazon, and others.
manoaction
Well-Known Member
As long as you have something, I'd consider running a teaspoon of the sediment through another starter. It should behave similar to a normal starter as in it should turn cloudy and then start to settle within three or four days. If you behavior takes weeks or if it smells wretched on the second starter, then I would suspect you don't have what your looking for.
dinnerstick
Well-Known Member
Ok after about 1.5 weeks of capturing some local bugs I have a fairly thick layer on the bottom of my mason jar pretty comprable to washing yeast. The smell is not at all unpleasant it almost smells like plain yogurt very lactic and somewhat sweet smelling initially somewhat honey like though there was only Dme and water. What I found interesting was that there were no bubbles like in a regular fermentation, does this mean I didn't catch any yeast that all i got was bacteria ? should I put it back outside ?
i have exactly the same thing. mine never bubbled or got foamy, then started to smell cheezy, now after 3 weeks i have convinced myself it smells sour/spicy. gf thinks it smells like cold vomit. i keep flipping between, "i've done something really disgusting here" and "total breakthrough!". i'm going to make a very small starter and add a little bit. all suggestions will be considered! i will also streak some out on plates and have a good hard stare at them
Pith
Well-Known Member
I can't wait to start doing this again. I just moved out of home, and now I have to go and get some DME...
manoaction
Well-Known Member
After stepping it up in a second starter, my homegrown guys are going to town in a one gallon test batch brewed with my backyard Chinook hops. The scent on the yeast starter was lemony/pineapple. We'll see how this goes...
bluedubbed
Active Member
This thread is awesome. I have an experiment on going and will post photos soon, but I had a question.
I have a few strains of wild yeast that metabolize bromocresol green - so they are white/opaque when grown on BCG, yet they are sensitive to cyclohexamide at 10-20 mg/L.
From what I've read, Brett should be resistant to cyclohexamide. Any ideas to what these yeast might be. They are clearly budding yeast.
Thanks.
Doing some more searching, it seems possible that they might be Candida? That would suck as all my isolates have the same phenotype. One more edit. I don't think many Candida can tolerate cycloheximide.
Another update. I actually got one clone after 3 days at 30 degrees C that grew on cyclohexamide (last picture). It's looks no different than (colony morphology or cells) the cyc negative yeast.
I grew small cultures (10 mL in DME with a small bit of yeast extract) and neither of the cultures have much flavor at all? Do I need larger/longer grows? Yes, I might need to grow the yeast on cycloheximide for 7-10 days.
Ok, here are some photos of two wild yeast clones isolated from different sources.
What do you guys think? I think they might be Brett.

Clone 1

Clone 1

cyclohexamide resistant Clone 2
I have a few strains of wild yeast that metabolize bromocresol green - so they are white/opaque when grown on BCG, yet they are sensitive to cyclohexamide at 10-20 mg/L.
From what I've read, Brett should be resistant to cyclohexamide. Any ideas to what these yeast might be. They are clearly budding yeast.
Thanks.
Doing some more searching, it seems possible that they might be Candida? That would suck as all my isolates have the same phenotype. One more edit. I don't think many Candida can tolerate cycloheximide.
Another update. I actually got one clone after 3 days at 30 degrees C that grew on cyclohexamide (last picture). It's looks no different than (colony morphology or cells) the cyc negative yeast.
I grew small cultures (10 mL in DME with a small bit of yeast extract) and neither of the cultures have much flavor at all? Do I need larger/longer grows? Yes, I might need to grow the yeast on cycloheximide for 7-10 days.
Ok, here are some photos of two wild yeast clones isolated from different sources.
What do you guys think? I think they might be Brett.

Clone 1

Clone 1

cyclohexamide resistant Clone 2
dinnerstick
Well-Known Member
can you send samples to a yeast lab for genotyping? i am very familiar with plant genotyping techniques and molecular biology in general, it's relatively simple to amplify a bit of ribosomal RNA genes or microsatellite DNA and sequence it, so i would imagine that a well equipped lab that already knows which bits to sequence could knock them out easily and give you a genus?? but i know nothing about yeast genotyping. if you are buying in bulk the total cost of reagents+materials to sequence a gene from one colony on a plate is probably around 20 dollars with the economy of scale (and if you happen to own a dye-terminator capillary sequencer!) and can be done from the amount of cells on the end of a toothpick stuck into the colony.
my point is it might not cost that much if there is a willing lab locally
my point is it might not cost that much if there is a willing lab locally
That really brings up a great point.... Is there a lab that anyone knows of to perform this identification. As well to do this service for under $1,000's of dollars. I would love to identify what I have caught as I will be slanting and isolating colonies.
That really brings up a great point.... Is there a lab that anyone knows of to perform this identification. As well to do this service for under $1,000's of dollars. I would love to identify what I have caught as I will be slanting and isolating colonies.
Siebel does it for 200 dollar or so. All they do is just a simple PCR of a specific part of the genome. I do not have the info here, but will post it next week. I tried the ITS parts myself as well, but those are hard to sequence because of the repeats. They use a different sequence.
I made a protocol where you can just use some yeast and PCR on them without doing DNA isolations. Yeast is a pain to lyse, but it worked. Then sequencing of the PCRed DNA is easy. A simple blast will give you your results.
Siebel does it for 200 dollar or so. All they do is just a simple PCR of a specific part of the genome. I do not have the info here, but will post it next week. I tried the ITS parts myself as well, but those are hard to sequence because of the repeats. They use a different sequence.
I made a protocol where you can just use some yeast and PCR on them without doing DNA isolations. Yeast is a pain to lyse, but it worked. Then sequencing of the PCRed DNA is easy. A simple blast will give you your results.
That would be great if you post that info it would be much appreciated.

dinnerstick
Well-Known Member
from the PLoS paper that someone just posted in this very forum
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0035507
For amplification of the ITS1/ITS4 domain of yeast 26S rDNA genes (ITS-TRFLP) [5], the forward primer used was ITS1HEX (5′-[5HEX] TCCGTAGGTGAACCTGCGG-3′ and the reverse primer was ITS4 (5′-TCCTCCGCTTATTGATATGC-3′
and the reverse primer was ITS4 (5′-TCCTCCGCTTATTGATATGC-3′ [6]. The PCR conditions were an initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 2 min, and with a final extension at 72°C for 7 min.
[6]. The PCR conditions were an initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 2 min, and with a final extension at 72°C for 7 min.
not even degenerate primers, so i guess they anneal to sacc, brett, candida, etc? haven't read the paper yet though, just the abstract and tiny bit of methods. tired. tomorrow.
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0035507
For amplification of the ITS1/ITS4 domain of yeast 26S rDNA genes (ITS-TRFLP) [5], the forward primer used was ITS1HEX (5′-[5HEX] TCCGTAGGTGAACCTGCGG-3′
not even degenerate primers, so i guess they anneal to sacc, brett, candida, etc? haven't read the paper yet though, just the abstract and tiny bit of methods. tired. tomorrow.
dinnerstick
Well-Known Member
sorry the protocol described above is for population TRFLP not direct sequencing from individual colonies
bluedubbed
Active Member
Yeah, there are some simple PCR assays that will distinguish Brett from others. As posted above and this paper as well - http://www.ncbi.nlm.nih.gov/pubmed?term=Tessonnière h
Also, this lab will do identification. Not sure of costs.
http://www.etslabs.com/
The cyc resistant clone in my last post is definitely Brett. I grew a large liquid culture and it is extremely sour.
The cyc sensitive clone is probably not a Brett. The culture smells sweet. Maybe a wild SC.
I'll post more once I get a small batch fermenting.
Also, this lab will do identification. Not sure of costs.
http://www.etslabs.com/
The cyc resistant clone in my last post is definitely Brett. I grew a large liquid culture and it is extremely sour.
The cyc sensitive clone is probably not a Brett. The culture smells sweet. Maybe a wild SC.
I'll post more once I get a small batch fermenting.
http://jaapie.org/?page_id=497 Here is a small write up of my methods. Siebel does it for 220 dollar.
For Siebel the species of the sample is determined by sequencing of the domain D1-
D2 of the 26s rDNA. The results were analysed using “Blast” database
(http://blast.ncbi.nlm.nih.gov/Blast.cgi).
The rDNA evolves slowly, therefore these primers will anneal to different species with reasonable efficiency.
For Siebel the species of the sample is determined by sequencing of the domain D1-
D2 of the 26s rDNA. The results were analysed using “Blast” database
(http://blast.ncbi.nlm.nih.gov/Blast.cgi).
The rDNA evolves slowly, therefore these primers will anneal to different species with reasonable efficiency.
bluedubbed
Active Member
jaapie- Thanks for the link to your post.
I isolated 10 more cycloheximide resistant clones in the mountains this weekend.
Time to start seeing what kind (hopefully different) flavors they produce.
Should be interesting.
I isolated 10 more cycloheximide resistant clones in the mountains this weekend.
Time to start seeing what kind (hopefully different) flavors they produce.
Should be interesting.
roymeo
Member
White Labs has various kits and lab services:
http://www.whitelabs.com/beer/craft_test_kits.html
http://www.whitelabs.com/beer/craft_lab.html
I noticed some info in the microscope bit about wild yeasts being much smaller than regular beer yeast strains.
roymeo
http://www.whitelabs.com/beer/craft_test_kits.html
http://www.whitelabs.com/beer/craft_lab.html
I noticed some info in the microscope bit about wild yeasts being much smaller than regular beer yeast strains.
roymeo
dinnerstick
Well-Known Member
jaapie- nice blog. love the DIC pics of yeast cells. i work at wageningen uni!
i'm not quite to the point of trying to genotype anything yet, but curious if anyone has an opinion from their experience, my wild ferment went from cheesy smelling to eventually distinctly sour and sort of phenolic, and of course like feet. it never bubbled, foamed, or moved the airlock. i didn't check the gravity before or after but it started around ~1.035. i was going to pitch some into an aerated starter to see what happened, but when i had a look at it the other day for the first time in 10 days or so there were clumps of yuckies, they looked like fuzzy raisins sort of. i gave it a swirl, took a bit out, did 6 serial 10-fold dilutions, plated them out. waiting to see what grows. i chucked the rest as i don't want to propagate the fuzzballs. i admit i don't know what i'm hoping for (or what i'm doing!). my original goal was just to make a spontaneously fermented sour but now i may as well see if there is something interesting in there to culture? could there be an appreciable sacc or brett culture going in there so slowly that i wouldn't have noticed it at work?
i'm not quite to the point of trying to genotype anything yet, but curious if anyone has an opinion from their experience, my wild ferment went from cheesy smelling to eventually distinctly sour and sort of phenolic, and of course like feet. it never bubbled, foamed, or moved the airlock. i didn't check the gravity before or after but it started around ~1.035. i was going to pitch some into an aerated starter to see what happened, but when i had a look at it the other day for the first time in 10 days or so there were clumps of yuckies, they looked like fuzzy raisins sort of. i gave it a swirl, took a bit out, did 6 serial 10-fold dilutions, plated them out. waiting to see what grows. i chucked the rest as i don't want to propagate the fuzzballs. i admit i don't know what i'm hoping for (or what i'm doing!). my original goal was just to make a spontaneously fermented sour but now i may as well see if there is something interesting in there to culture? could there be an appreciable sacc or brett culture going in there so slowly that i wouldn't have noticed it at work?
jaapie- nice blog. love the DIC pics of yeast cells. i work at wageningen uni!
i'm not quite to the point of trying to genotype anything yet, but curious if anyone has an opinion from their experience, my wild ferment went from cheesy smelling to eventually distinctly sour and sort of phenolic, and of course like feet. it never bubbled, foamed, or moved the airlock. i didn't check the gravity before or after but it started around ~1.035. i was going to pitch some into an aerated starter to see what happened, but when i had a look at it the other day for the first time in 10 days or so there were clumps of yuckies, they looked like fuzzy raisins sort of. i gave it a swirl, took a bit out, did 6 serial 10-fold dilutions, plated them out. waiting to see what grows. i chucked the rest as i don't want to propagate the fuzzballs. i admit i don't know what i'm hoping for (or what i'm doing!). my original goal was just to make a spontaneously fermented sour but now i may as well see if there is something interesting in there to culture? could there be an appreciable sacc or brett culture going in there so slowly that i wouldn't have noticed it at work?
Really, good old Wageningen! Man, I miss that town. Lived there for over 10 years. Worked at Microbiology when they were still behind the old Unitas building. Now I live in suburbia USA, what a bummer that is for a change. Anyways!
Hm, you should have checked the gravity that would have helped. The thing is, with these wild ferments, you cannot really hurry those, nor tell where you will end up. You have to really wait it out for not just months, but even years. It might well be that it needs more time to get high enough numbers of your organisms you want to see in there to start fermenting. Spontaneous fermented beers also contain a ****-ton of bacteria, and can look/smell really disgusting for the first year.
dinnerstick
Well-Known Member
i work at the fancy new campus, but live in utrecht center. couldn't face life in the sticks! so i can't say i know the town well, just started here after working at utrecht uni for a while.
i can handle the smell, and i am prepared to wait out a spontaneous fermentation, i have other sour / sourish (from wyeast cultures) beers aging, but i quit the game on this experiment when i saw brown balls floating in there. is it possible that moldy (was that mold?) balls can be part of the succession of microfauna? this wort was just some extract+tiny bit of hop left out overnight, rather than a 'real' beer, if it looked like it was on the right track i was going to knock out a lambic style wort and inoculate with this stuff, and check back in a year. the thinking that i was at a disadvantage not brewing in a brewery / barrels with an enormous population of the right critters resident to kick things off before any mold could take hold. back to the drawing board though i guess, or should i just put all my chips on one square, make the beer, subject it to my terrace coolship (pot) treatment, let it go? hmmm..
i can handle the smell, and i am prepared to wait out a spontaneous fermentation, i have other sour / sourish (from wyeast cultures) beers aging, but i quit the game on this experiment when i saw brown balls floating in there. is it possible that moldy (was that mold?) balls can be part of the succession of microfauna? this wort was just some extract+tiny bit of hop left out overnight, rather than a 'real' beer, if it looked like it was on the right track i was going to knock out a lambic style wort and inoculate with this stuff, and check back in a year. the thinking that i was at a disadvantage not brewing in a brewery / barrels with an enormous population of the right critters resident to kick things off before any mold could take hold. back to the drawing board though i guess, or should i just put all my chips on one square, make the beer, subject it to my terrace coolship (pot) treatment, let it go? hmmm..
dinnerstick
Well-Known Member
these are my plates, anyone have a guess from colony morphology? 36 hours on YDP plates at room temp ~21. each chamber is a 0.1 dilution of the previous, first is 0.1 dilution of the 'beer'. to my untrained eye (i grow a couple types of bacteria and very occasionally laboratory sacc at work) it loos like only 2 types of colony, bigger creamy yeasty one and smaller shiny bacterial(?) one?? i expected a lot more crazy diversity! will give the plates a couple more days
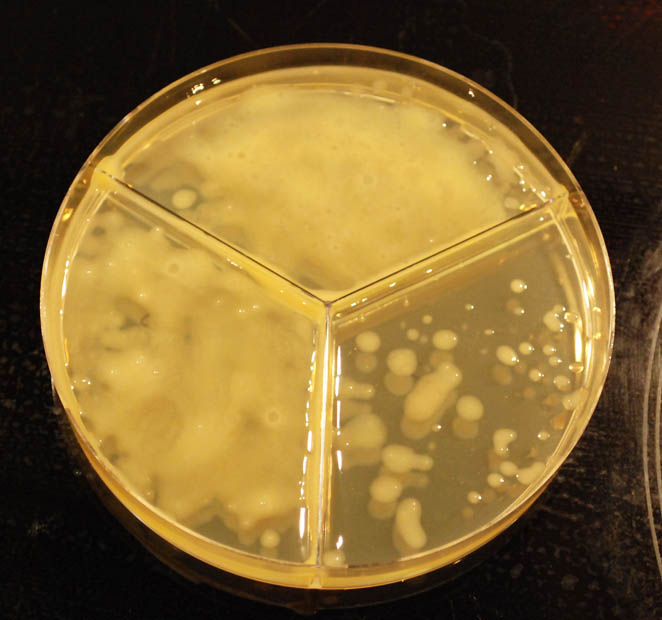
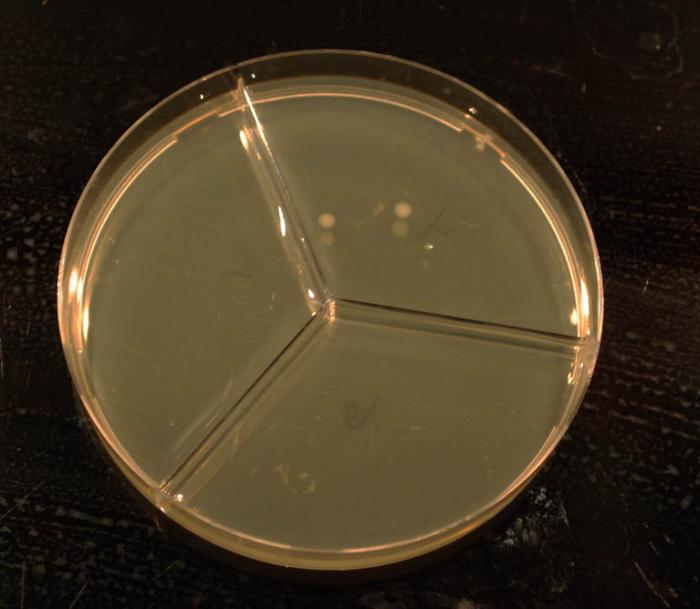


bluedubbed
Active Member
dinnerstick- Not sure based on morphology, but I found that plates with chloramphenicol (25ug/ml) were really helpful in getting rid of bacterial contamination and cycloheximinde (10-20 ug/ml) were useful for enriching for potential Brett. I've started making plates with both when I try to pull out new isolates. Seems like you might have access to these. I found brocresol green to be less useful.
dinnerstick
Well-Known Member
bluedubbed, thanks i have chlor and cx. i guess i could streak some of the lawn of the high concentrated chambers onto those plates. meanwhile i stuck one of the creamy colonies under the scope out of curiosity and saw these buggers, didn't get a scale bar on there but that's almost the full field of view with a 40x objective. sorry for the poor quality.
they looked like miniature tictacs on crack

they looked like miniature tictacs on crack

dinnerstick
Well-Known Member
That gooey stuff is probably bacteria. I have seen that too. Simple test would be a microscope. You will know immediately.
wow good timing, as i was uploading the pic! yep bacteria
bluedubbed
Active Member
Another question.
I have over 10 different pure Brett cultures (or at least I think so) isolated at various times and places over many hundreds of miles. While I bet most are the same, I thought I might get a few different species or even sub types based on the different time and location of sampling. When I culture them in WLN or YPD media, they really all seem to smell/taste similar to each other. Does anyone know if the different Brett species or sub types have distinctive metabolites that can be detected in this manner, i.e., growing in a yeast media and smelling/tasting? Or should I be doing this experiment with small batches of wort? I might go ahead with 2 of the faster growing isolates and brew a few batches.
Thanks for the help.
I have over 10 different pure Brett cultures (or at least I think so) isolated at various times and places over many hundreds of miles. While I bet most are the same, I thought I might get a few different species or even sub types based on the different time and location of sampling. When I culture them in WLN or YPD media, they really all seem to smell/taste similar to each other. Does anyone know if the different Brett species or sub types have distinctive metabolites that can be detected in this manner, i.e., growing in a yeast media and smelling/tasting? Or should I be doing this experiment with small batches of wort? I might go ahead with 2 of the faster growing isolates and brew a few batches.
Thanks for the help.
I got some pictures of wild yeast trap in the barn:

I can't tell if that's a krausen or a pellicle. Either way, I'm very excited. I tried to poke my finger through it, and it actually coated my finger enough to keep the beer off of it! I tilted a little wort into my hand, and it was a bit sticky but it wasn't sour at all.


I can't tell if that's a krausen or a pellicle. Either way, I'm very excited. I tried to poke my finger through it, and it actually coated my finger enough to keep the beer off of it! I tilted a little wort into my hand, and it was a bit sticky but it wasn't sour at all.

By cell photos, doesn't look as rodlike as Bret. If sampled at temp higher than 70f could be cryptoccocus. I'm not savy on pcr, but you could maybe look at fatty acid profile by a hexane extraction and then gc analysis of fatty acid profile. I believe the crypto will have more lipids that are of a unique variety compared to sacch. For reference, I wash wild samples using a trap to catch the early foam, then make a starter from that foam with a 8 brix soln. Then I sample from here and play. With this method I've been able to sustain some poly cultures of Bret and sacch. If you want to clean up some bacteria, you might also experiment with some ph and alcohol concentration variations. Lowering the ph and increasing the alcohol content worked to clean up several samples. Plating individual colonies to Identify by pcr is a noble cause. Please post pcr method when you find it.
biodavid said:Here is a Bret sacch polyculture at 40x
This was sampled from the skin of an overripe nectarine that was hanging from a tree and appeared to be a fermentation in progress.
By cell photos, doesn't look as rodlike as Bret. If sampled at temp higher than 70f could be cryptoccocus. I'm not savy on pcr, but you could maybe look at fatty acid profile by a hexane extraction and then gc analysis of fatty acid profile. I believe the crypto will have more lipids that are of a unique variety compared to sacch. For reference, I wash wild samples using a trap to catch the early foam, then make a starter from that foam with a 8 brix soln. Then I sample from here and play. With this method I've been able to sustain some poly cultures of Bret and sacch. If you want to clean up some bacteria, you might also experiment with some ph and alcohol concentration variations. Lowering the ph and increasing the alcohol content worked to clean up several samples. Plating individual colonies to Identify by pcr is a noble cause. Please post pcr method when you find it.
Brett is not always rods, they often have lemon shaped cells as well. I have a few brett strains and they are not rod shaped. The PCR method was just posted a few posts ago
I got some pictures of wild yeast trap in the barn:

I can't tell if that's a krausen or a pellicle. Either way, I'm very excited. I tried to poke my finger through it, and it actually coated my finger enough to keep the beer off of it! I tilted a little wort into my hand, and it was a bit sticky but it wasn't sour at all.

Looks very nice. I have cultured wild yeasts, and fermented spontaneous, but have only in a few occasions seen a pellicle.
I noticed some info in the microscope bit about wild yeasts being much smaller than regular beer yeast strains.
roymeo
that is because polyploidy is more common in beer yeasts. The cells become larger when there are more copies of the genome present. Actually interesting, the same effect happens in vegetables on a multi-level scale (think about the beefsteak tomato).
dinnerstick
Well-Known Member
thanks everyone for the great advice on this thread. i'm going back to the drawing board and will re-emerge at a later time!
with yeast
with yeast
bluedubbed
Active Member
Got 2 new isolates that smell super funky when grown in WLN media. I'm going to do a few mini batches this weekend with 3 different isolates and see what flavors/aromas these guys produce with barley! I'll update in a few weeks. Such a different morphology between these two isolates. Photos taken at same magnification.
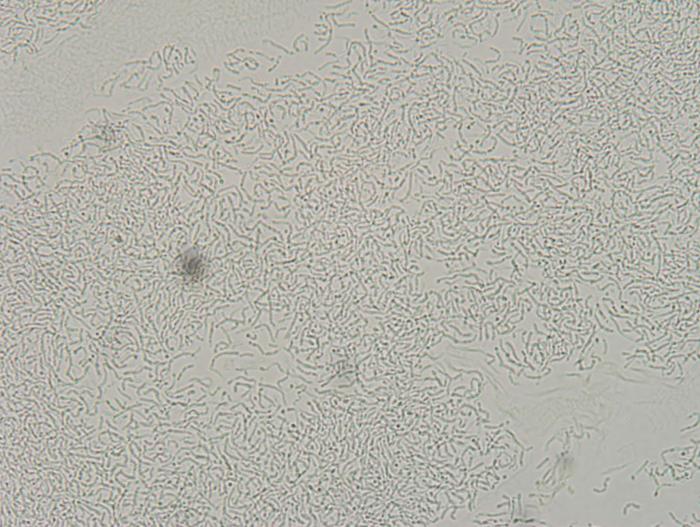
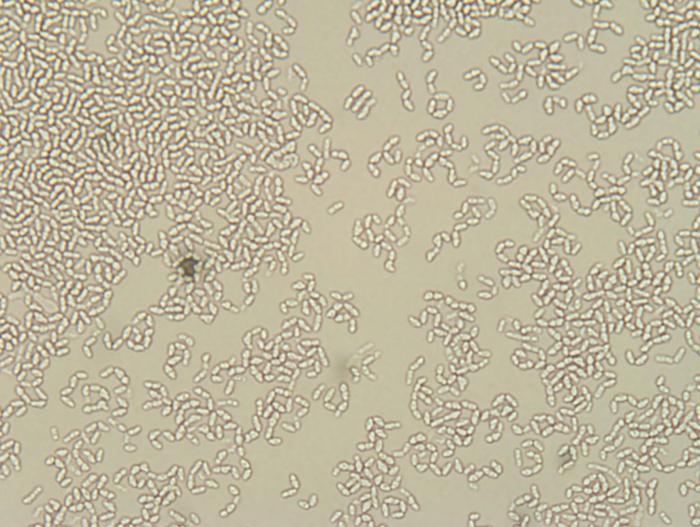


bluedubbed
Active Member
The photos are at 64X. They should be pure since the grows were from single colonies after a few rounds of selection. I'll take a few photos of the plates, but surprisingly, colony morphology doesn't seem all that different between the different isolates I have. That said, I'm not a yeast geneticist so I'm sure I'm missing subtle differences.
The photos are at 64X. They should be pure since the grows were from single colonies after a few rounds of selection. I'll take a few photos of the plates, but surprisingly, colony morphology doesn't seem all that different between the different isolates I have. That said, I'm not a yeast geneticist so I'm sure I'm missing subtle differences.
You could let the colonies go for two weeks, and see how they look when they are giant.
thasnazzle
Well-Known Member
Has anyone had any experience trying to capture wild yeast with aged hops? I just got a lb of aged hops in and I thought it might help the wild yeast capture process... am I correct on that?
Has anyone had any experience trying to capture wild yeast with aged hops? I just got a lb of aged hops in and I thought it might help the wild yeast capture process... am I correct on that?
hello
no this is not correct.
hops does not help capture wild yeast.
in lambic beers, overaged hops are used because they still have their bacteriostatic power without their original bittering features.
wathever you do, you need to use hops in any wort, specifically if you want to inoculate with wild yeasts as the process might be long. This for the same bacteriostatic reason.
in a nutshell, if you have overaged hops around, you sure can use them for this purpose!
Similar threads
- Replies
- 10
- Views
- 865


